Embryo Image Analysis
My team’s goal was to train a neural network that would be able to recognize and measure the Trophectoderm Epithelium (TE) and Inner Cell Mass (ICM) of an embryo with a high accuracy. This network would then be able to be used in the process of choosing embryos for use in in vitro fertilization (IVF). If the network was accurate, Embryologists could use it to measure the ICM and TE of potential embryos that were going to be used in IVF instead of doing it visually which can lead to human error due to the reliance on personal judgment. The network helps in grading the potential embryos so the chance of a successful IVF procedure is greater.
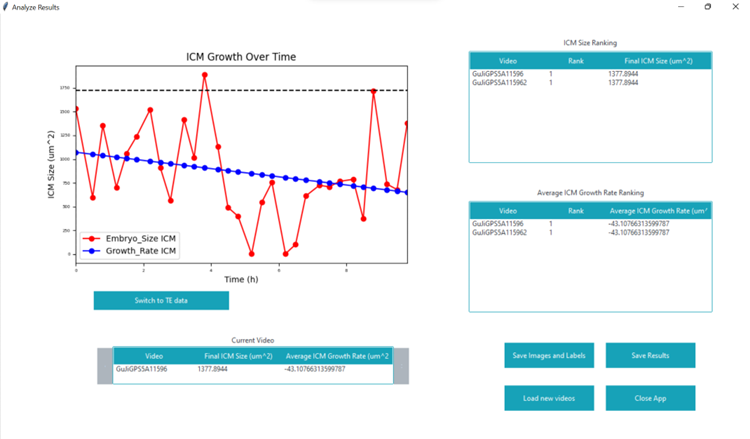
Since we were given a relatively small amount of images labeled with the TE and ICM by embryologists, my job consisted of augmenting these images so that we could train the neural networks with more images. I developed code for a brightness augmentation that changes the brightness of the images so the network could be trained with images of many different brightnesses. Other augmentations done by previous members were mixed with the brightness augmentation and our job was to find a combination of augmentations that yielded the highest accuracy. I also worked on the graphical user interface (GUI) that displayed the results of the neural network. This GUI used the Tkinter library of Python.
From this project I developed much stronger abilities in Python coding and working in teams on a major project.
Some of my work on the GUI can be seen in this picture: